Histone deacetylase 5
| HDAC5 | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||||||||||
| Aliases | HDAC5, HD5, NY-CO-9, histone deacetylase 5 | ||||||||||||||||||||||||
| External IDs | MGI: 1333784 HomoloGene: 3995 GeneCards: HDAC5 | ||||||||||||||||||||||||
| |||||||||||||||||||||||||
| |||||||||||||||||||||||||
| |||||||||||||||||||||||||
| |||||||||||||||||||||||||
| Orthologs | |||||||||||||||||||||||||
| Species | Human | Mouse | |||||||||||||||||||||||
| Entrez |
|
| |||||||||||||||||||||||
| Ensembl |
|
| |||||||||||||||||||||||
| UniProt |
|
| |||||||||||||||||||||||
| RefSeq (mRNA) |
|
| |||||||||||||||||||||||
| RefSeq (protein) |
|
| |||||||||||||||||||||||
| Location (UCSC) | Chr 17: 44.08 – 44.12 Mb | Chr 11: 102.19 – 102.23 Mb | |||||||||||||||||||||||
PubMed search | [3] | [4] | |||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||
| |||||||||||||||||||||||||
Histone deacetylase 5 is an enzyme that in humans is encoded by the HDAC5 gene.[5][6][7]
Contents
1 Function
2 Interactions
3 See also
4 References
5 Further reading
6 External links
Function
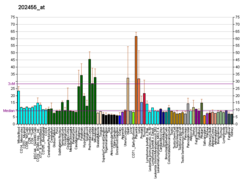
Histones play a critical role in transcriptional regulation, cell cycle progression, and developmental events. Histone acetylation/deacetylation alters chromosome structure and affects transcription factor access to DNA. The protein encoded by this gene belongs to the class II histone deacetylase/acuc/apha family. It possesses histone deacetylase activity and represses transcription when tethered to a promoter. It coimmunoprecipitates only with HDAC3 family member and might form multicomplex proteins. It also interacts with myocyte enhancer factor-2 (MEF2) proteins, resulting in repression of MEF2-dependent genes. This gene is thought to be associated with colon cancer. Two transcript variants encoding different isoforms have been found for this gene.[7]
AMP-activated protein kinase regulation of the glucose transporter GLUT4 occurs by phosphorylation of HDAC5.[8]
HDAC5 is involved in memory consolidation and suggests that development of more selective HDAC inhibitors for the treatment of Alzheimer's disease should avoid targeting HDAC5.[9] Its function can be effectively examined by siRNA knockdown based on an independent validation.[10]
Interactions
Histone deacetylase 5 has been shown to interact with:
BCL6,[11]
CBX5,[12]
GATA1,[13]
HDAC3,[5][14][15][16]
IKZF1,[17]
MEF2A,[18]
NRIP1,[19]
NCOR1,[14][20]
NCOR2,[20]
YWHAQ,[21] and
ZBTB16.[11][22]
See also
- Histone deacetylase
References
^ abc GRCh38: Ensembl release 89: ENSG00000108840 - Ensembl, May 2017
^ abc GRCm38: Ensembl release 89: ENSMUSG00000008855 - Ensembl, May 2017
^ "Human PubMed Reference:"..mw-parser-output cite.citationfont-style:inherit.mw-parser-output .citation qquotes:"""""""'""'".mw-parser-output .citation .cs1-lock-free abackground:url("//upload.wikimedia.org/wikipedia/commons/thumb/6/65/Lock-green.svg/9px-Lock-green.svg.png")no-repeat;background-position:right .1em center.mw-parser-output .citation .cs1-lock-limited a,.mw-parser-output .citation .cs1-lock-registration abackground:url("//upload.wikimedia.org/wikipedia/commons/thumb/d/d6/Lock-gray-alt-2.svg/9px-Lock-gray-alt-2.svg.png")no-repeat;background-position:right .1em center.mw-parser-output .citation .cs1-lock-subscription abackground:url("//upload.wikimedia.org/wikipedia/commons/thumb/a/aa/Lock-red-alt-2.svg/9px-Lock-red-alt-2.svg.png")no-repeat;background-position:right .1em center.mw-parser-output .cs1-subscription,.mw-parser-output .cs1-registrationcolor:#555.mw-parser-output .cs1-subscription span,.mw-parser-output .cs1-registration spanborder-bottom:1px dotted;cursor:help.mw-parser-output .cs1-ws-icon abackground:url("//upload.wikimedia.org/wikipedia/commons/thumb/4/4c/Wikisource-logo.svg/12px-Wikisource-logo.svg.png")no-repeat;background-position:right .1em center.mw-parser-output code.cs1-codecolor:inherit;background:inherit;border:inherit;padding:inherit.mw-parser-output .cs1-hidden-errordisplay:none;font-size:100%.mw-parser-output .cs1-visible-errorfont-size:100%.mw-parser-output .cs1-maintdisplay:none;color:#33aa33;margin-left:0.3em.mw-parser-output .cs1-subscription,.mw-parser-output .cs1-registration,.mw-parser-output .cs1-formatfont-size:95%.mw-parser-output .cs1-kern-left,.mw-parser-output .cs1-kern-wl-leftpadding-left:0.2em.mw-parser-output .cs1-kern-right,.mw-parser-output .cs1-kern-wl-rightpadding-right:0.2em
^ "Mouse PubMed Reference:".
^ ab Grozinger CM, Hassig CA, Schreiber SL (June 1999). "Three proteins define a class of human histone deacetylases related to yeast Hda1p". Proc Natl Acad Sci U S A. 96 (9): 4868–73. doi:10.1073/pnas.96.9.4868. PMC 21783. PMID 10220385.
^ Scanlan MJ, Chen YT, Williamson B, Gure AO, Stockert E, Gordan JD, Tureci O, Sahin U, Pfreundschuh M, Old LJ (June 1998). "Characterization of human colon cancer antigens recognized by autologous antibodies". Int J Cancer. 76 (5): 652–8. doi:10.1002/(SICI)1097-0215(19980529)76:5<652::AID-IJC7>3.0.CO;2-P. PMID 9610721.
^ ab "Entrez Gene: HDAC5 histone deacetylase 5".
^ McGee SL, van Denderen BJ, Howlett KF, Mollica J, Schertzer JD, Kemp BE, Hargreaves M (2008). "AMP-activated protein kinase regulates GLUT4 transcription by phosphorylating histone deacetylase 5". Diabetes. 57 (4): 860–867. doi:10.2337/db07-0843. PMID 18184930.
^ Agis-Balboa RC, Pavelka Z, Kerimoglu C, Fischer A (January 2013). "Loss of HDAC5 impairs memory function: implications for Alzheimer's disease". J Alzheimers Dis. 33 (1): 35–44. doi:10.3233/JAD-2012-121009. PMID 22914591.
^ Munkácsy, Gyöngyi; Sztupinszki, Zsófia; Herman, Péter; Bán, Bence; Pénzváltó, Zsófia; Szarvas, Nóra; Győrffy, Balázs (2016-01-01). "Validation of RNAi Silencing Efficiency Using Gene Array Data shows 18.5% Failure Rate across 429 Independent Experiments". Molecular Therapy - Nucleic Acids. 5: e366. doi:10.1038/mtna.2016.66. ISSN 2162-2531. PMC 5056990. PMID 27673562.
^ ab Lemercier C, Brocard MP, Puvion-Dutilleul F, Kao HY, Albagli O, Khochbin S (2002). "Class II histone deacetylases are directly recruited by BCL6 transcriptional repressor". J. Biol. Chem. 277 (24): 22045–52. doi:10.1074/jbc.M201736200. PMID 11929873.
^ Zhang CL, McKinsey TA, Olson EN (2002). "Association of class II histone deacetylases with heterochromatin protein 1: potential role for histone methylation in control of muscle differentiation". Mol. Cell. Biol. 22 (20): 7302–12. doi:10.1128/MCB.22.20.7302-7312.2002. PMC 139799. PMID 12242305.
^ Watamoto K, Towatari M, Ozawa Y, Miyata Y, Okamoto M, Abe A, Naoe T, Saito H (2003). "Altered interaction of HDAC5 with GATA-1 during MEL cell differentiation". Oncogene. 22 (57): 9176–84. doi:10.1038/sj.onc.1206902. PMID 14668799.
^ ab Zhang J, Kalkum M, Chait BT, Roeder RG (2002). "The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2". Mol. Cell. 9 (3): 611–23. doi:10.1016/S1097-2765(02)00468-9. PMID 11931768.
^ Fischle W, Dequiedt F, Hendzel MJ, Guenther MG, Lazar MA, Voelter W, Verdin E (2002). "Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR". Mol. Cell. 9 (1): 45–57. doi:10.1016/S1097-2765(01)00429-4. PMID 11804585.
^ Grozinger CM, Schreiber SL (2000). "Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization". Proc. Natl. Acad. Sci. U.S.A. 97 (14): 7835–40. doi:10.1073/pnas.140199597. PMC 16631. PMID 10869435.
^ Koipally J, Georgopoulos K (2002). "A molecular dissection of the repression circuitry of Ikaros". J. Biol. Chem. 277 (31): 27697–705. doi:10.1074/jbc.M201694200. PMID 12015313.
^ Lemercier C, Verdel A, Galloo B, Curtet S, Brocard MP, Khochbin S (2000). "mHDA1/HDAC5 histone deacetylase interacts with and represses MEF2A transcriptional activity". J. Biol. Chem. 275 (20): 15594–9. doi:10.1074/jbc.M908437199. PMID 10748098.
^ Castet A, Boulahtouf A, Versini G, Bonnet S, Augereau P, Vignon F, Khochbin S, Jalaguier S, Cavaillès V (2004). "Multiple domains of the Receptor-Interacting Protein 140 contribute to transcription inhibition". Nucleic Acids Res. 32 (6): 1957–66. doi:10.1093/nar/gkh524. PMC 390375. PMID 15060175.
^ ab Huang EY, Zhang J, Miska EA, Guenther MG, Kouzarides T, Lazar MA (2000). "Nuclear receptor corepressors partner with class II histone deacetylases in a Sin3-independent repression pathway". Genes Dev. 14 (1): 45–54. PMC 316335. PMID 10640275.
^ Vega RB, Harrison BC, Meadows E, Roberts CR, Papst PJ, Olson EN, McKinsey TA (2004). "Protein kinases C and D mediate agonist-dependent cardiac hypertrophy through nuclear export of histone deacetylase 5". Mol. Cell. Biol. 24 (19): 8374–85. doi:10.1128/MCB.24.19.8374-8385.2004. PMC 516754. PMID 15367659.
^ Chauchereau A, Mathieu M, de Saintignon J, Ferreira R, Pritchard LL, Mishal Z, Dejean A, Harel-Bellan A (2004). "HDAC4 mediates transcriptional repression by the acute promyelocytic leukaemia-associated protein PLZF". Oncogene. 23 (54): 8777–84. doi:10.1038/sj.onc.1208128. PMID 15467736.
Further reading
.mw-parser-output .refbeginfont-size:90%;margin-bottom:0.5em.mw-parser-output .refbegin-hanging-indents>ullist-style-type:none;margin-left:0.mw-parser-output .refbegin-hanging-indents>ul>li,.mw-parser-output .refbegin-hanging-indents>dl>ddmargin-left:0;padding-left:3.2em;text-indent:-3.2em;list-style:none.mw-parser-output .refbegin-100font-size:100%
Verdin E, Dequiedt F, Kasler HG (2003). "Class II histone deacetylases: versatile regulators". Trends Genet. 19 (5): 286–93. doi:10.1016/S0168-9525(03)00073-8. PMID 12711221.
Huang EY, Zhang J, Miska EA, et al. (2000). "Nuclear receptor corepressors partner with class II histone deacetylases in a Sin3-independent repression pathway". Genes Dev. 14 (1): 45–54. PMC 316335. PMID 10640275.
Lemercier C, Verdel A, Galloo B, et al. (2000). "mHDA1/HDAC5 histone deacetylase interacts with and represses MEF2A transcriptional activity". J. Biol. Chem. 275 (20): 15594–9. doi:10.1074/jbc.M908437199. PMID 10748098.
Grozinger CM, Schreiber SL (2000). "Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3- 3-dependent cellular localization". Proc. Natl. Acad. Sci. U.S.A. 97 (14): 7835–40. doi:10.1073/pnas.140199597. PMC 16631. PMID 10869435.
Huynh KD, Fischle W, Verdin E, Bardwell VJ (2000). "BCoR, a novel corepressor involved in BCL-6 repression". Genes Dev. 14 (14): 1810–23. doi:10.1101/gad.14.14.1810 (inactive 2019-02-19). PMC 316791. PMID 10898795.
Mahlknecht U, Schnittger S, Ottmann OG, et al. (2000). "Chromosomal organization and localization of the human histone deacetylase 5 gene (HDAC5)". Biochim. Biophys. Acta. 1493 (3): 342–8. doi:10.1016/S0167-4781(00)00191-3. PMID 11018260.
Zhang CL, McKinsey TA, Lu JR, Olson EN (2001). "Association of COOH-terminal-binding protein (CtBP) and MEF2-interacting transcription repressor (MITR) contributes to transcriptional repression of the MEF2 transcription factor". J. Biol. Chem. 276 (1): 35–9. doi:10.1074/jbc.M007364200. PMID 11022042.
McKinsey TA, Zhang CL, Lu J, Olson EN (2000). "Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation". Nature. 408 (6808): 106–11. doi:10.1038/35040593. PMC 4459600. PMID 11081517.
McKinsey TA, Zhang CL, Olson EN (2001). "Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14-3-3 to histone deacetylase 5". Proc. Natl. Acad. Sci. U.S.A. 97 (26): 14400–5. doi:10.1073/pnas.260501497. PMC 18930. PMID 11114197.
Fischle W, Dequiedt F, Fillion M, et al. (2001). "Human HDAC7 histone deacetylase activity is associated with HDAC3 in vivo". J. Biol. Chem. 276 (38): 35826–35. doi:10.1074/jbc.M104935200. PMID 11466315.
McKinsey TA, Zhang CL, Olson EN (2001). "Identification of a Signal-Responsive Nuclear Export Sequence in Class II Histone Deacetylases". Mol. Cell. Biol. 21 (18): 6312–21. doi:10.1128/MCB.21.18.6312-6321.2001. PMC 87361. PMID 11509672.
Ozawa Y, Towatari M, Tsuzuki S, et al. (2001). "Histone deacetylase 3 associates with and represses the transcription factor GATA-2". Blood. 98 (7): 2116–23. doi:10.1182/blood.V98.7.2116. PMID 11567998.
Potter GB, Beaudoin GM, DeRenzo CL, et al. (2001). "The hairless gene mutated in congenital hair loss disorders encodes a novel nuclear receptor corepressor". Genes Dev. 15 (20): 2687–701. doi:10.1101/gad.916701. PMC 312820. PMID 11641275.
Fischle W, Dequiedt F, Hendzel MJ, et al. (2002). "Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR". Mol. Cell. 9 (1): 45–57. doi:10.1016/S1097-2765(01)00429-4. PMID 11804585.
Lemercier C, Brocard MP, Puvion-Dutilleul F, et al. (2002). "Class II histone deacetylases are directly recruited by BCL6 transcriptional repressor". J. Biol. Chem. 277 (24): 22045–52. doi:10.1074/jbc.M201736200. PMID 11929873.
Huang Y, Tan M, Gosink M, et al. (2002). "Histone deacetylase 5 is not a p53 target gene, but its overexpression inhibits tumor cell growth and induces apoptosis". Cancer Res. 62 (10): 2913–22. PMID 12019172.
External links
HDAC5+protein,+human at the US National Library of Medicine Medical Subject Headings (MeSH)
This article incorporates text from the United States National Library of Medicine, which is in the public domain.